SAS to R Migration: How to Import, Process, and Export SAS Files in R
<p><b>UPDATED: </b>April, 2025.</p>
<p>When it comes to data analytics, it's no secret that R gives you way more bang for your buck than when compared to SAS.</p>
<p>R won't cost you a penny, but the same can't be said for SAS licenses. In addition, R gives you access to thousands of packages created by other data scientists who've probably already solved the exact problem you're dealing with. Plus, when you need help, there's a huge community ready to jump in with answers on places like Stack Overflow or GitHub.</p>
<p><b>Why does this matter to you?</b> If you're working at a company that's been using SAS forever but wants to modernize, you can't just flip a switch overnight. You need both systems to play nice during the transition, letting you leverage your existing SAS files while building new workflows in R.</p>
<blockquote><a href="https://www.appsilon.com/post/sas-vs-r-programming" target="_blank">SAS vs R Programming: Which to Choose and How to Switch</a></blockquote>
<p>In this article, we'll show you exactly how to read, manipulate, and write SAS files in R so you can seamlessly bridge these two worlds.</p>
<h3>Table of contents</h3>
- <a href="#introduction">Introduction to SAS</a>
- <a href="#read">How to Read SAS Files in R</a>
- <a href="#write">How to Save R DataFrames to SAS Files</a>
- <a href="#example">Example: Aggregate SAS File in R</a>
- <a href="#best-practices">R and SAS Best Practices</a>
- <a href="#summary">Summing up R and SAS</a>
<h2 id="introduction">Introduction to SAS</h2>
<p>SAS (Statistical Analysis System) has been the go-to analytics software for many large enterprises and government agencies since the 1970s. It's especially common in regulated industries like healthcare, finance, and pharmaceuticals where reputation for stability and validation matters the most.</p>
<p><b>This doesn't mean large companies aren't transitioning to open-source.</b> Read this recent example:</p>
<ul>
<li><a href="https://www.appsilon.com/post/gsk-r-adoption-journey" target="_blank">GSK's Open-Source Shift: Training 1,000 Biostatisticians in R</a></li>
</ul>
<p><b>But what exactly makes SAS different from R?</b> The biggest distinction is that SAS is a commercial, proprietary software with a focus on enterprise-level data processing. You'll find it's great at handling massive datasets and comes with excellent technical support — if you can afford the hefty price tag.</p>
<p>Many organizations have built their entire data infrastructure around SAS. This means there's tons of legacy data stored in SAS formats that you'll need to access even as you transition to R. That's why knowing how to work with SAS files is an essential skill in your data science toolkit.</p>
<blockquote><a href="https://www.appsilon.com/post/r-vs-sas-pharma-life-sciences">Beyond SAS: How R is Revolutionizing Pharma and Life Sciences</a></blockquote>
<h3>Different SAS file types</h3>
<p>Before we dive into the code, let's understand what we're working with. SAS uses a couple of different file formats, but these are the two you'll encounter most often:</p>
<ul>
<li><b>SAS Data Files (<code>.sas7bdat</code>)</b> - These are the bread and butter of SAS, similar to data frames in R. They store tabular data with rows and columns, including numeric, character, and date variables. Think of them as Excel.</li>
<li><b>SAS Transport Files (<code>.xpt</code>)</b> - These are platform-independent files designed for sharing SAS datasets between different systems. They're handy when you need to transfer data between SAS and other software like R.</li>
</ul>
<p>There are other SAS file types too, like catalog files (<code>.sas7bcat</code>) that store metadata, but you'll be dealing with .sas7bdat and .xpt files most of the time when working between SAS and R.</p>
<p>Now that you understand what you're working with, let's look at how to actually read these files in R.</p>
<blockquote><a href="https://www.appsilon.com/post/open-source-pharma" target="_blank">Open-Source Adoption in Pharma: Opportunities and Challenges</a></blockquote>
<h2 id="read">How to Read SAS Files in R</h2>
<p>Reading SAS files in R is easy thanks to the <code>{haven}</code> package, part of the <a href="http://tidyverse.org" target="_blank">tidyverse</a> ecosystem. First, you'll need to install it if you haven't already:</p>
<pre><code class="language-r">install.packages("haven")</code></pre>
<p>Once installed, load the package and use the <code>read_sas()</code> function to import your SAS data file:</p>
<pre><code class="language-r">library(haven)
# Dataset: https://github.com/RhoInc/sas-codebook/blob/master/ExampleFiles/cars.sas7bdat
sas_data <- read_sas("data/cars.sas7bdat")
head(sas_data)</code></pre>
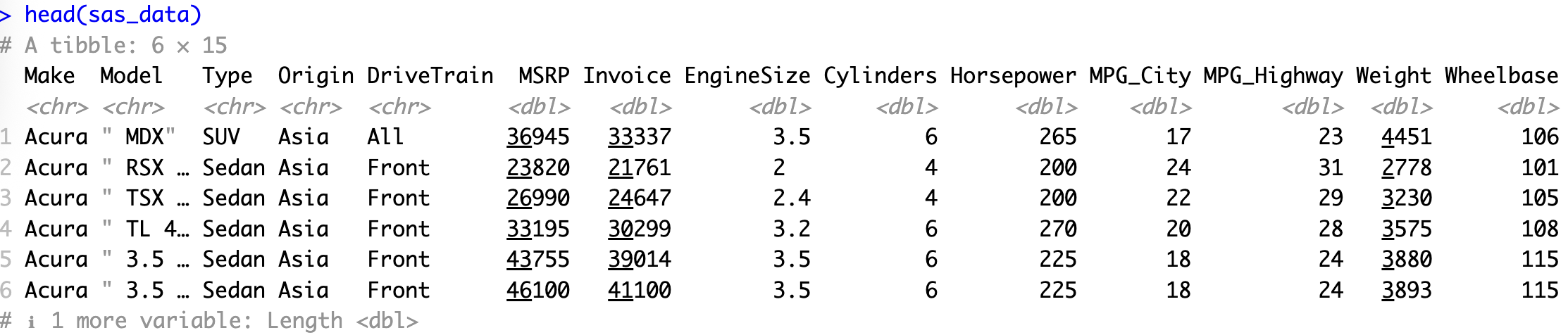
<p>That's it! Your SAS dataset is now available as an R data frame (technically, a tibble). The <code>head()</code> function gives you a quick peek at the first few rows of your imported data.</p>
<p>For this example, we've used a <a href="https://github.com/RhoInc/sas-codebook/blob/master/ExampleFiles/cars.sas7bdat" target="_blank">cars dataset that's freely available on GitHub</a>. Feel free to download it if you want to follow along with the code.</p>
<h3>Encoding issues</h3>
<p>Sometimes you'll run into encoding problems when importing SAS files, especially if they contain special characters or were created in a different locale. If you see weird symbols where characters should be, try specifying the encoding:</p>
<pre><code class="language-r">read_sas("data/cars.sas7bdat", encoding = "UTF-8")</code></pre>

<p>Common encodings include "UTF-8" (the most universal), "latin1" (common in Western Europe), and "windows-1252" (common on Windows systems). You might need to try a few different options depending on how your SAS file was created.</p>
<h3>Dealing with SAS labels</h3>
<p>One of SAS's distinctive features is its labeling system. SAS uses labels to attach metadata to values, which is different from how R handles factors. The <code>haven</code> package provides a special function called "labelled" to help bridge this gap.</p>
<p>Here's a simple example of how labelled vectors work in R:</p>
<pre><code class="language-r">x1 <- labelled(
sample(1:5),
c(Good = 1, Bad = 5)
)
x2 <- labelled(
c("M", "F", "F", "F", "M"),
c(Male = "M", Female = "F")
)</code></pre>

<p>You can then include these labelled vectors in a data frame (or tibble):</p>
<pre><code class="language-r">library(tibble)
tibble(x1, x2, z = 1:5)</code></pre>

<p>When <code>haven</code> imports a SAS dataset with labels, it automatically converts them to this labelled class. This preserves the original information while making it accessible in R.</p>
<h2 id="write">How to Save R DataFrames to SAS Files</h2>
<p>Once you've done your data manipulation in R, you'll often need to send the results back to colleagues who are still using SAS. The <code>haven</code> package makes this possible, but there's a crucial distinction you need to understand first.</p>
<p><b>Here's a critical warning about exporting to SAS:</b> Even though <code>haven</code> offers two export functions, only one of them actually works reliably. The <code>write_sas()</code> function attempts to create native <code>.sas7bdat</code> files, but since the SAS format is proprietary and poorly documented, these files often can't be opened by SAS itself. Always use <code>write_xpt()</code> instead, which creates SAS transport files (<code>.xpt</code>) that SAS can consistently read without issues.</p>
<p>In other words, if you want your SAS colleagues to actually be able to open your files, use <code>write_xpt()</code> instead of <code>write_sas()</code>. Here's how to do it:</p>
<pre><code class="language-r">my_data <- data.frame(
ID = 1:5,
Name = c("Bob", "Ed", "Rod", "Dav", "Eva"),
Value = c(90, 85, 78, 92, 88)
)
write_xpt(
my_data,
path = "data/output_file.xpt"
)
# Confirm
read_xpt("data/output_file.xpt")</code></pre>
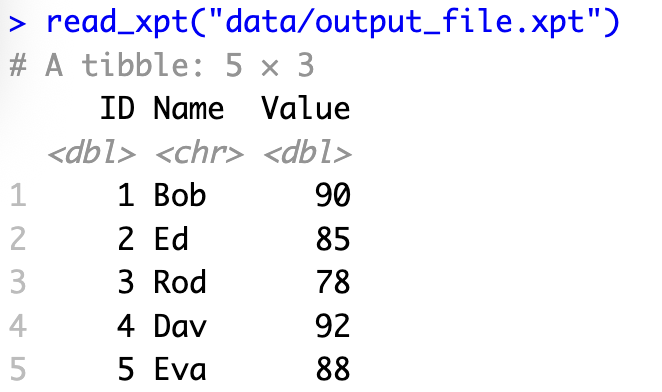
<p>The code above creates a simple data frame with three columns and then exports it as a SAS transport file. The last line reads the file back in to confirm that the export worked correctly.</p>
<h3>Handle missing values</h3>
<p>R and SAS handle missing values differently. In R, we use <code>NA</code>, while SAS has multiple types of missing values. The <code>haven</code> package lets you work with these differences through the <code>tagged_na()</code> function:</p>
<pre><code class="language-r">my_data <- data.frame(
ID = 1:5,
Name = c("Bob", "Ed", "Rod", "Dav", "Eva"),
Value = c(90, 85, 78, 92, 88),
na_values = tagged_na("Not applicable")
)
write_xpt(
my_data,
path = "data/output_file_with_na.xpt"
)
# Confirm
read_xpt("data/output_file_with_na.xpt")</code></pre>
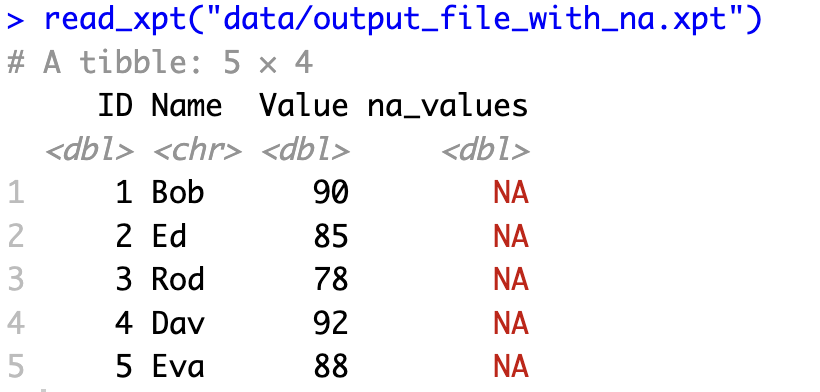
<p>This approach lets you preserve the meaning behind different types of missing values when moving between R and SAS. For example, you might want to distinguish between "Not applicable," "Refused to answer," and "Don't know" in survey data.</p>
<p>When you read this file back into R, you'll see the tagged NA values preserving the original information. Similarly, when your SAS colleagues open the file, they'll see the appropriate missing value codes.</p>
<p>Now that you know how to both read and write SAS files in R, let's put these skills together in a practical example.</p>
<h2 id="example">Example: Aggregate SAS File in R</h2>
<p>Let's put everything together with a practical example. One common task when transitioning from SAS to R is recreating your SAS summary reports in R and then sharing the results back with your SAS-using colleagues.</p>
<p>We'll use the cars dataset again, group it by vehicle type, and calculate summary statistics - similar to what you might do with <code>PROC MEANS</code> in SAS.</p>
<p>First, let's import the data and create a summary report with the summarytools package:</p>
<pre><code class="language-r">library(dplyr)
library(summarytools)
data <- read_sas("data/cars.sas7bdat")
grouped_data <- data %>%
group_by(Type)
view(dfSummary(grouped_data))</code></pre>
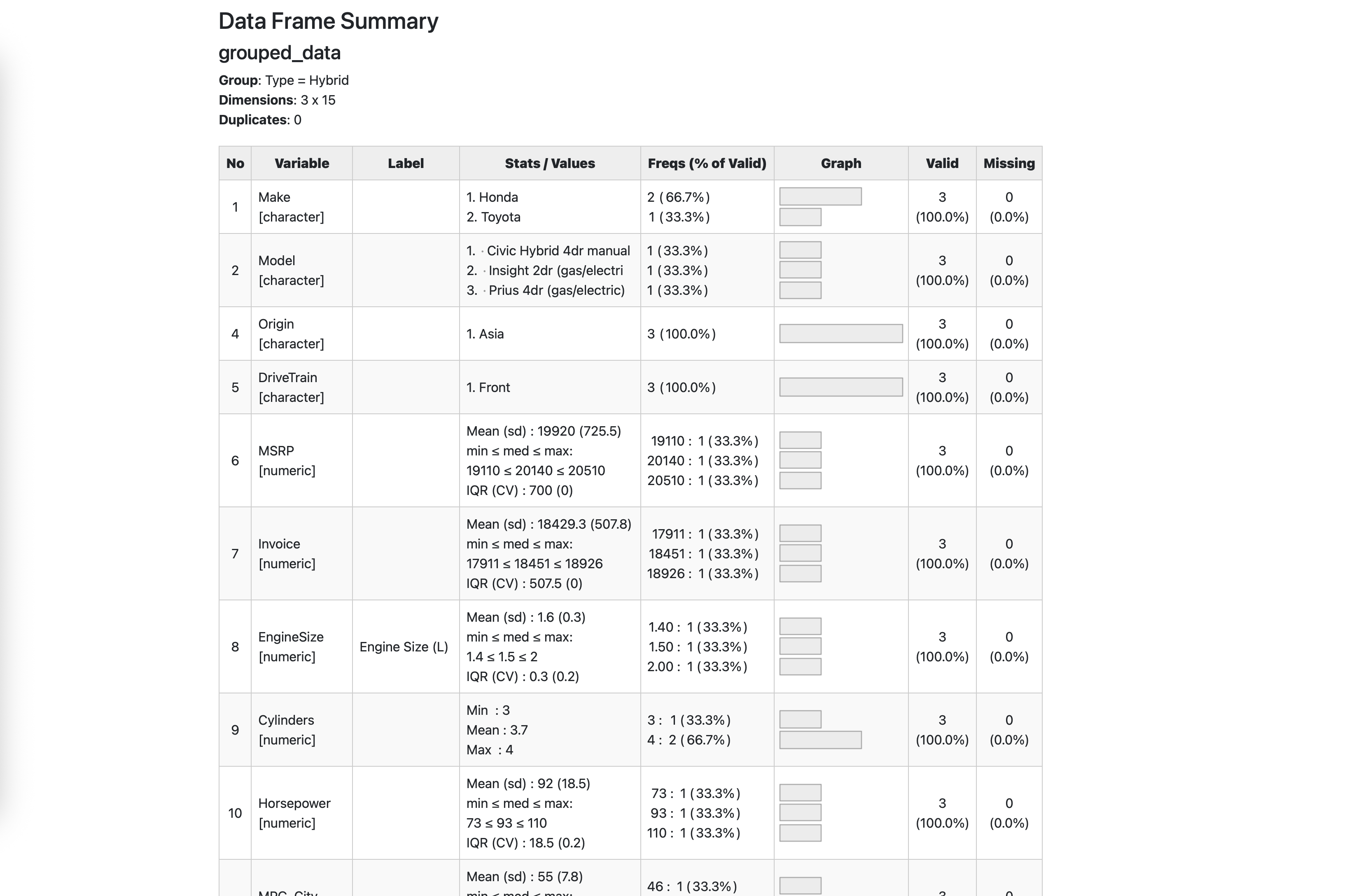
<p>The code above will open an HTML report in your viewer pane and show detailed statistics for each variable in the dataset, grouped by the vehicle type. In other words, ut's a quick way to get a comprehensive overview of your data.</p>
<p>But what if you want more control over exactly which statistics to calculate? Let's create a custom summary and export it back to SAS format:</p>
<pre><code class="language-r">summarised_data <- data %>%
group_by(Type) %>%
summarise(
across(
where(is.numeric),
list(
mean = mean,
stdev = sd,
median = median,
min = min,
max = max,
iqr = ~IQR(..1, na.rm = TRUE)
)
)
)
summarised_data
write_xpt(
summarised_data,
path = "data/cars_summarised.xpt"
)</code></pre>

<p>This more advanced example uses dplyr's powerful <code>across()</code> function to apply multiple summary statistics to every numeric column in our dataset. Here's what's happening:</p>
<ul>
<li>We group the data by vehicle type.</li>
<li>We use <code>summarise()</code> with <code>across()</code> to apply functions to multiple columns at once.</li>
<li><code>where(is.numeric)</code> selects only the numeric columns.</li>
<li>We apply six different statistics to each numeric column.</li>
<li>Finally, we export the result as a SAS transport file.</li>
</ul>
<p>The result is a summary table with columns named like "MPG_City_mean", "MPG_City_median", etc. This gives you a similar output to what you'd get from PROC MEANS in SAS, but with the flexibility and power of R's data manipulation tools.</p>
<p>If your SAS colleagues need this summary data for further analysis or reporting, they can import the <code>.xpt</code> file using standard SAS code like:</p>
<pre><code class="language-sas">libname mylib xport "path/to/cars_summarised.xpt";
data work.cars_summary;
set mylib.summarised_data;
run;</code></pre>
<p>And that's all there is to it!</p>
<h2 id="best-practices">R and SAS Best Practices</h2>
<p>Transitioning from SAS to R won't always be smooth sailing. Hopefully, you can minimize risks by following these best practices:</p>
<ul>
<li><b>Document everything: </b>Use R Markdown or Quarto to create reproducible reports that track how you processed the data. This is crucial when transitioning between systems and helps both you and your colleagues understand each transformation step.</li>
<li><b>Handle missing values properly: </b>R and SAS treat missing values differently. Be explicit about how you want to handle NAs when moving between systems, especially if certain missing values have specific meanings in your data.</li>
<li><b>Test your workflow: </b>Always verify that your exported SAS files can be read correctly by having a SAS user try to open and use them. This simple step can save hours of troubleshooting later.</li>
<li><b>Seek help:</b> If you require further guidance, don't hesitate to seek help from the <a href="https://forum.posit.co" target="_blank">Posit community</a> or <a href="https://communities.sas.com/" target="_blank">SAS communities</a>. Collaboration can often lead to quicker solutions.</li>
</ul>
<h2 id="summary">Summing up R and SAS</h2>
<p>We've covered a lot of ground in this guide to working with SAS files in R. You now know how to import SAS datasets into R, process them, and export the results back to SAS format for your colleagues.</p>
<p>The <code>haven</code> package makes this cross-platform workflow possible, and it gives you the freedom to use R's flexibility while still collaborating with teams that rely on SAS. Remember to use <code>read_sas()</code> for importing and <code>write_xpt()</code> (not <code>write_sas()</code>) for exporting to ensure maximum compatibility.</p>
<p>If you're in the middle of a SAS to R migration at your organization, this hybrid approach lets you gradually transition workflows without disrupting existing processes. You can start building new analysis in R while maintaining compatibility with legacy SAS systems.</p>
<p>Want to get more out of your data with custom analytics solutions? We're here to help you modernize your data workflow. Whether you're just starting your SAS to R migration or looking to enhance your existing R capabilities, our team of experts can guide you through the process.</p>
<blockquote><b><a href="https://www.appsilon.com/contact-us" target="_blank">Reach out to Appsilon to transition from SAS to R</a></b></blockquote>

